Welcome to CWAS-Plus documentation!
CWAS-Plus (Category-Wide Association Study) is a data analysis tool designed to conduct rigorous association tests for discovering noncoding associations in complex genomic disorders. It runs category-based burden tests using variants from whole-genome sequencing data and various annotation datasets. CWAS-Plus provides a user-friendly interface for efficient hypothesis testing and has promising implications for uncovering the pathophysiology of various genomic disorders.
Here are the reference papers:
An analytical framework for whole genome sequence association studies and its implications for autism spectrum disorder (Werling et al., 2018)
Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder (An et al., 2018)
CWAS-Plus: Estimating genome-wide evaluation of noncoding variation from whole genome sequencing data. (Kim et al., in preperation)
CWAS-Plus workflow
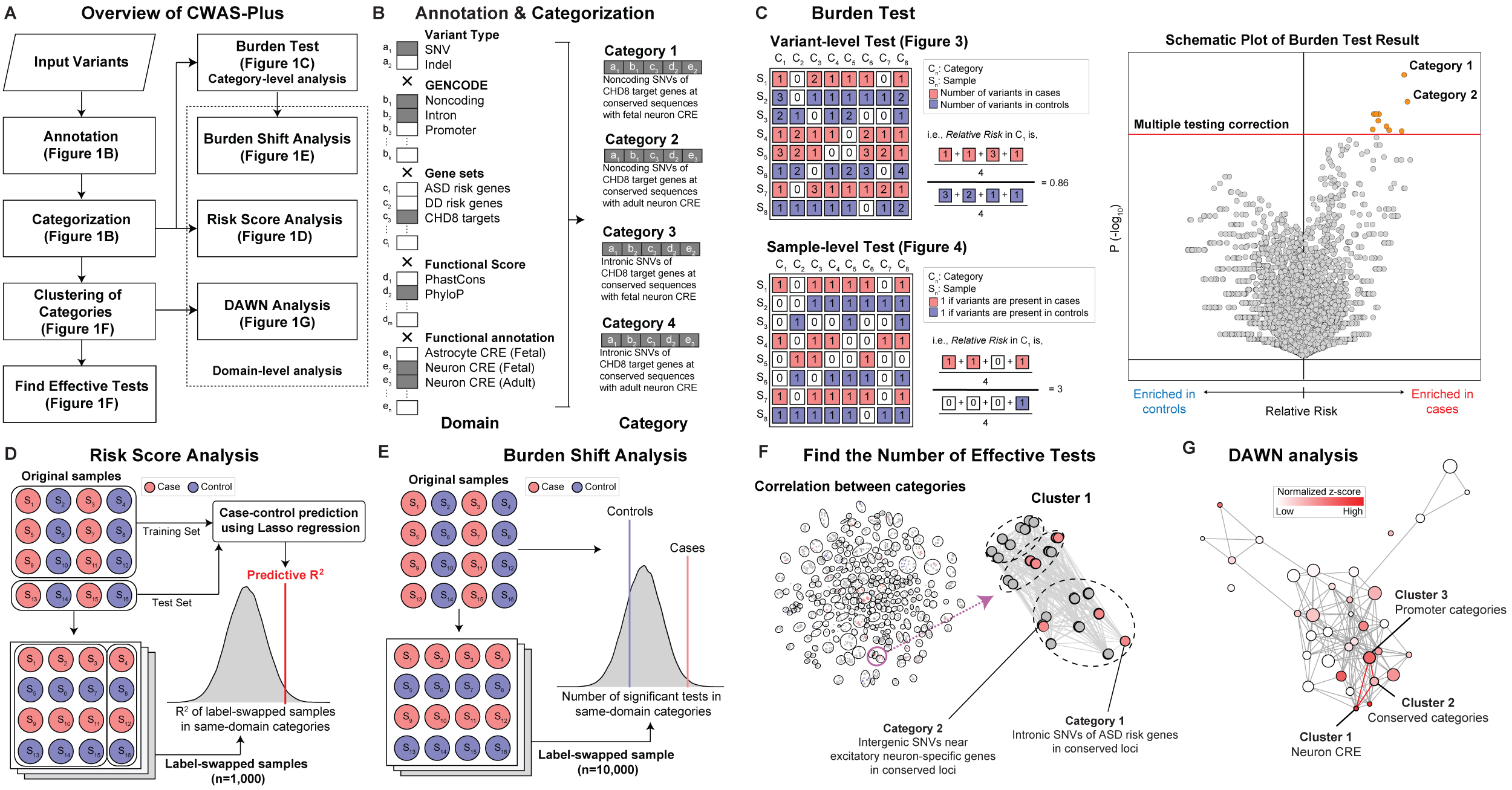
A. Workflow: Variants extracted from whole-genome sequencing data of samples (sample variant) serve as inputs. The three steps depicted in the black box correspond to the final outputs obtained from CWAS-Plus. B-G. Graphic descriptions of each process in CWAS-Plus: Red (case) and blue (control) represent the phenotype. F. Representative network pointed by the purple arrow provides an enlarged view of a subset of the network. The color indicates the direction of the burden in each category (red, case burden; blue, control burden). G. The color of the clusters reflects the scale of the normalized z-score, which represents the degree of disease association of the cluster. Darker red indicates a higher association. Circle size represents the number of categories within the cluster.
CWAS-Plus tutorial
CWAS-Plus installation
CWAS-Plus requirements
CWAS-Plus configuration
CWAS-Plus steps
Other useful commands
Misc.
Here is the original CWAS repository: sanderslab/cwas